# LucaPCycle
We developed a dual-channel model named LucaPCycle, based on the raw sequence and protein language large models, to predict whether a protein sequence has phosphate-solubilizing functionality and its specific type among the 31 fine-grained functions.
We constructed two models, including an identification model(binary classification) and a fine-grained classification of specific phosphate-solubilizing functional types(31 classification).
## 1. Model Architecture
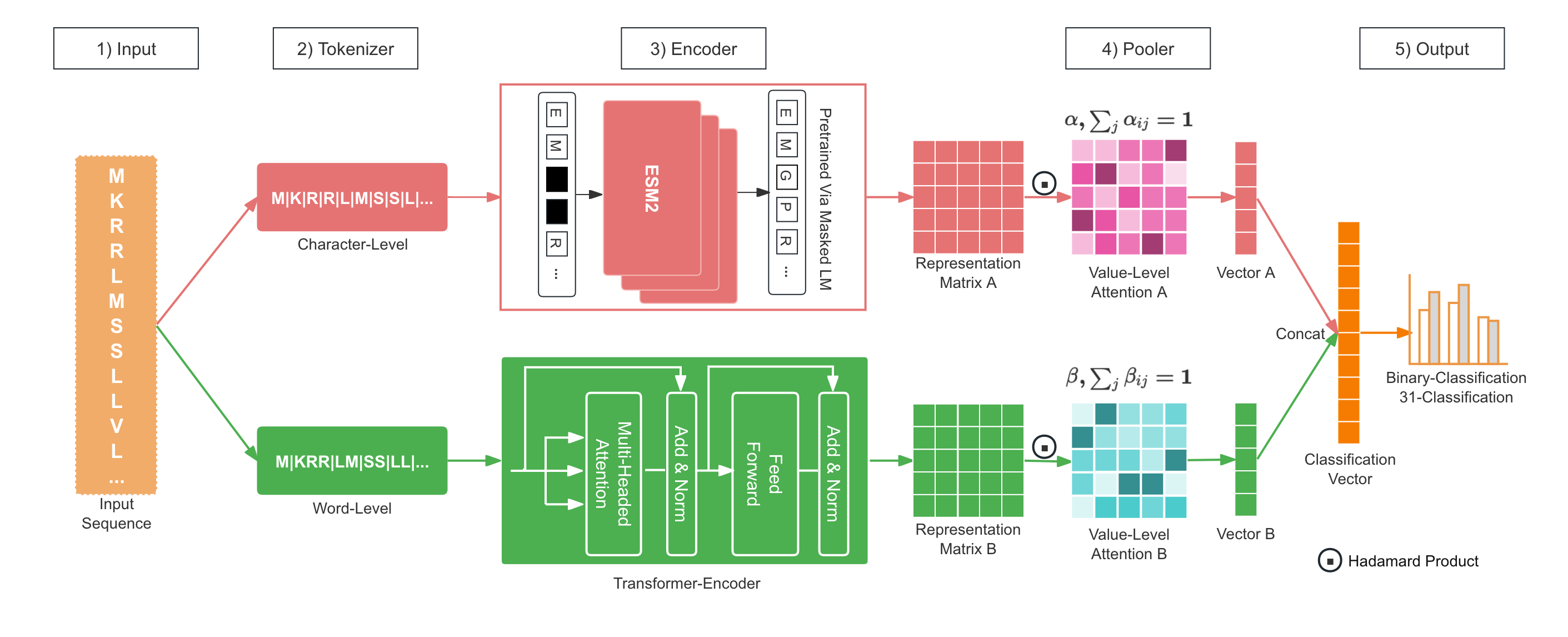 **Fig.1 LucaPCycle.**
## 2. Environment Installation
### step1: update git
#### 1) centos
sudo yum update
sudo yum install git-all
#### 2) ubuntu
sudo apt-get update
sudo apt install git-all
### step2: install python 3.9
#### 1) download anaconda3
wget https://repo.anaconda.com/archive/Anaconda3-2022.05-Linux-x86_64.sh
#### 2) install conda
sh Anaconda3-2022.05-Linux-x86_64.sh
##### Notice: Select Yes to update ~/.bashrc
source ~/.bashrc
#### 3) create a virtual environment: python=3.9.13
conda create -n lucapcycle python=3.9.13
#### 4) activate lucapcycle
conda activate lucapcycle
### step3: install other requirements
pip install -r requirements.txt -i https://pypi.tuna.tsinghua.edu.cn/simple
## 3. Inference
### TrainedCheckPoint
Trained LucaPCycle Checkpoint FTP: TrainedCheckPoint for LucaPCycle
**Notice**
The project will download automatically LucaPCycle Trained-CheckPoint from **FTP**.
When downloading automatically failed, you can manually download:
Copy the **TrainedCheckPoint Files(`models/` + `logs/`)** from http://47.93.21.181/lucapcycle/TrainedCheckPoint/* into the project.
### Usage
Firstly, predict whether a sequence has phosphate-solubilizing functionality.
The inference script: **`src/prediction.py`** or **`src/prediction.sh`**
```python prediction.py -h``` for **help**
#### Binary Classification
```shell
cd src/
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python prediction.py \
--fasta ../test_data/examples.fasta \
--llm_truncation_seq_length 4096 \
--model_path .. \
--save_path ../predicted_results/test_data/examples_predicted.csv \
--dataset_name extra_p_2_class_v2 \
--dataset_type protein \
--task_type binary_class \
--task_level_type seq_level \
--model_type lucaprot \
--input_type seq_matrix \
--time_str 20240120061735 \
--step 955872 \
--threshold 0.2 \
--per_num 1000 \
--gpu_id 0
```
#### 31 Classification
Then, for the sequences predicted to be positive in the **2-classification** inference, the fine-grained classification of specific phosphate-solubilizing functional types(**31 classes**) is further predicted.
```shell
cd src/
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python prediction.py \
--fasta ../test_data/example_positives.fasta \
--llm_truncation_seq_length 4096 \
--model_path .. \
--save_path ../predicted_results/test_data/example_positives_fine_grained_predicted.csv \
--dataset_name extra_p_31_class_v2 \
--dataset_type protein \
--task_type multi_class \
--task_level_type seq_level \
--model_type lucaprot \
--input_type seq_matrix \
--time_str 20240120061524 \
--step 294536 \
--per_num 1000 \
--gpu_id 1
```
#### Parameters
1) Input data parameters:
* fasta: `Path`, the input fasta filepath(for a batch samples)
* seq_id: `str`, the seq id(for one sample)
* seq: `str`, the sequence(for one sample)
* save_path: `Path`, the saved dir path of the batch samples predicted results(only for batch prediction)
2) Trained LucaPCycle checkpoint parameters:
* model_path: `Path`, model dir path,default: `../` (meaning the checkpoint in the project)
* dataset_name: `str`, the checkpoint version: `extra_p_2_class_v2`(2-classification) or `extra_p_31_class_v2`(31-classification)
* dataset_type: `str`, only `protein`, default: `protein`
* task_type: `str`, the trained task type: `binary_class`(2-classification) or `multi_class`(31-classification)
* task_level_type: `str`, sequence-level tasks, default: `seq-level`
* model_type: `str`, the model type, default: `lucaprot`
* input_type: `str`, the model channels, default: `seq_matrix`
* time_str: `str`, the trained checkpoint running time str: `20240120061735`(2-classification) or `20240120061524`(31-classification)
* step: `int`, the checkpoint step: `955872`(2-classification) or `294536`(31-classification)
3) Running parameters:
* topk: `int`, the topk labels when inferring 31-classification, default: `None`(meaining k=1)
* llm_truncation_seq_length: `int`, the max seq length to truncation(depends on the length of your sequence and the size of your GPU memory. default: `4096`
* per_num: `int`, the print progress is determined by how many sequences are predicted. default: `1000`
* threshold: `float`, the threshold for binary-classification, default: `0.1`, (positive>=threshold, negativeDataset FTP.
The raw data of LucaPCycle building in **`data/`** or Raw Data FTP,
where folder **`31P_genes/`** is fasta for each of the 31 fine-grained phosphate-solubilizing types,
and the file **`cold_spring_sample_50.csv`** is the non-redundancy sequences(including positives and negatives) using the CD-HIT tool with 50% sequence identity.
### 2) Large-scale Identification
The large-scale unidentified data is in **`inference_data/`** or Large-scale Unidentified Data FTP, total of **151,187,265** sequences.
The data includes 164 metagenomes and 33 metatranscriptomes,
which is sourced from sediment samples (sediment depths: 0-68.55 mbsf; water depths 860-3005 m) collected at 16 globally distributed cold seep sites.
These samples encompass five types of cold seeps, namely gas hydrates (n = 39), mud volcanoes (n = 7), asphalt volcanoes (n = 7), oil and gas seeps (n = 15) and methane seeps (n = 96).
The predicted results of the large-scale data are list in **`results/`** or Results FTP:
The file in the format of **`*_init*`** is the unchecked results, and the file in the format of **`*_verified*`** is the result of the verification by through three distinct methods: **ECOD Domain Analysis**, **DeepFRI v1.0.0 (Deep Functional Residue Identification)**, and **CLEAN v1.0.1 (Contrastive Learning-Enabled Enzyme Annotation)**.
* **LucaPCycle**
Results in **`results/LucaPCycle/`** or LucaPCycle Results FTP:
Resulting in **1,481,237** positive sequences.
The detailed predicted numbers for each class are shown below.
**Notice:** There may be **interesting findings**. Totaling 134,227 positive sequences(predicted by LucaPCycle) in file **`results/LucaPCycle/lucapcycle_unverifiable.fasta`** (9.06%) could not be confirmed using existing verified methods.
`lucapcycle_details_init.csv`: unchecked predicted details positives by LucaPCycle(include top1 prob and label, top10 prob and label)
`lucapcycle_init.ids.labels` & `lucapcycle_init.fasta`: unchecked predicted positives by LucaPCycle.
`lucapcycle_verified.ids.labels` & `lucapcycle_verified.fasta`: checked predicted positives by LucaPCycle.
`lucapcycle_unverifiable.ids` & `lucapcycle_unverifiable.ids`: unverifiable predicted positives by LucaPCycle.
**Fig.1 LucaPCycle.**
## 2. Environment Installation
### step1: update git
#### 1) centos
sudo yum update
sudo yum install git-all
#### 2) ubuntu
sudo apt-get update
sudo apt install git-all
### step2: install python 3.9
#### 1) download anaconda3
wget https://repo.anaconda.com/archive/Anaconda3-2022.05-Linux-x86_64.sh
#### 2) install conda
sh Anaconda3-2022.05-Linux-x86_64.sh
##### Notice: Select Yes to update ~/.bashrc
source ~/.bashrc
#### 3) create a virtual environment: python=3.9.13
conda create -n lucapcycle python=3.9.13
#### 4) activate lucapcycle
conda activate lucapcycle
### step3: install other requirements
pip install -r requirements.txt -i https://pypi.tuna.tsinghua.edu.cn/simple
## 3. Inference
### TrainedCheckPoint
Trained LucaPCycle Checkpoint FTP: TrainedCheckPoint for LucaPCycle
**Notice**
The project will download automatically LucaPCycle Trained-CheckPoint from **FTP**.
When downloading automatically failed, you can manually download:
Copy the **TrainedCheckPoint Files(`models/` + `logs/`)** from http://47.93.21.181/lucapcycle/TrainedCheckPoint/* into the project.
### Usage
Firstly, predict whether a sequence has phosphate-solubilizing functionality.
The inference script: **`src/prediction.py`** or **`src/prediction.sh`**
```python prediction.py -h``` for **help**
#### Binary Classification
```shell
cd src/
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python prediction.py \
--fasta ../test_data/examples.fasta \
--llm_truncation_seq_length 4096 \
--model_path .. \
--save_path ../predicted_results/test_data/examples_predicted.csv \
--dataset_name extra_p_2_class_v2 \
--dataset_type protein \
--task_type binary_class \
--task_level_type seq_level \
--model_type lucaprot \
--input_type seq_matrix \
--time_str 20240120061735 \
--step 955872 \
--threshold 0.2 \
--per_num 1000 \
--gpu_id 0
```
#### 31 Classification
Then, for the sequences predicted to be positive in the **2-classification** inference, the fine-grained classification of specific phosphate-solubilizing functional types(**31 classes**) is further predicted.
```shell
cd src/
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python prediction.py \
--fasta ../test_data/example_positives.fasta \
--llm_truncation_seq_length 4096 \
--model_path .. \
--save_path ../predicted_results/test_data/example_positives_fine_grained_predicted.csv \
--dataset_name extra_p_31_class_v2 \
--dataset_type protein \
--task_type multi_class \
--task_level_type seq_level \
--model_type lucaprot \
--input_type seq_matrix \
--time_str 20240120061524 \
--step 294536 \
--per_num 1000 \
--gpu_id 1
```
#### Parameters
1) Input data parameters:
* fasta: `Path`, the input fasta filepath(for a batch samples)
* seq_id: `str`, the seq id(for one sample)
* seq: `str`, the sequence(for one sample)
* save_path: `Path`, the saved dir path of the batch samples predicted results(only for batch prediction)
2) Trained LucaPCycle checkpoint parameters:
* model_path: `Path`, model dir path,default: `../` (meaning the checkpoint in the project)
* dataset_name: `str`, the checkpoint version: `extra_p_2_class_v2`(2-classification) or `extra_p_31_class_v2`(31-classification)
* dataset_type: `str`, only `protein`, default: `protein`
* task_type: `str`, the trained task type: `binary_class`(2-classification) or `multi_class`(31-classification)
* task_level_type: `str`, sequence-level tasks, default: `seq-level`
* model_type: `str`, the model type, default: `lucaprot`
* input_type: `str`, the model channels, default: `seq_matrix`
* time_str: `str`, the trained checkpoint running time str: `20240120061735`(2-classification) or `20240120061524`(31-classification)
* step: `int`, the checkpoint step: `955872`(2-classification) or `294536`(31-classification)
3) Running parameters:
* topk: `int`, the topk labels when inferring 31-classification, default: `None`(meaining k=1)
* llm_truncation_seq_length: `int`, the max seq length to truncation(depends on the length of your sequence and the size of your GPU memory. default: `4096`
* per_num: `int`, the print progress is determined by how many sequences are predicted. default: `1000`
* threshold: `float`, the threshold for binary-classification, default: `0.1`, (positive>=threshold, negativeDataset FTP.
The raw data of LucaPCycle building in **`data/`** or Raw Data FTP,
where folder **`31P_genes/`** is fasta for each of the 31 fine-grained phosphate-solubilizing types,
and the file **`cold_spring_sample_50.csv`** is the non-redundancy sequences(including positives and negatives) using the CD-HIT tool with 50% sequence identity.
### 2) Large-scale Identification
The large-scale unidentified data is in **`inference_data/`** or Large-scale Unidentified Data FTP, total of **151,187,265** sequences.
The data includes 164 metagenomes and 33 metatranscriptomes,
which is sourced from sediment samples (sediment depths: 0-68.55 mbsf; water depths 860-3005 m) collected at 16 globally distributed cold seep sites.
These samples encompass five types of cold seeps, namely gas hydrates (n = 39), mud volcanoes (n = 7), asphalt volcanoes (n = 7), oil and gas seeps (n = 15) and methane seeps (n = 96).
The predicted results of the large-scale data are list in **`results/`** or Results FTP:
The file in the format of **`*_init*`** is the unchecked results, and the file in the format of **`*_verified*`** is the result of the verification by through three distinct methods: **ECOD Domain Analysis**, **DeepFRI v1.0.0 (Deep Functional Residue Identification)**, and **CLEAN v1.0.1 (Contrastive Learning-Enabled Enzyme Annotation)**.
* **LucaPCycle**
Results in **`results/LucaPCycle/`** or LucaPCycle Results FTP:
Resulting in **1,481,237** positive sequences.
The detailed predicted numbers for each class are shown below.
**Notice:** There may be **interesting findings**. Totaling 134,227 positive sequences(predicted by LucaPCycle) in file **`results/LucaPCycle/lucapcycle_unverifiable.fasta`** (9.06%) could not be confirmed using existing verified methods.
`lucapcycle_details_init.csv`: unchecked predicted details positives by LucaPCycle(include top1 prob and label, top10 prob and label)
`lucapcycle_init.ids.labels` & `lucapcycle_init.fasta`: unchecked predicted positives by LucaPCycle.
`lucapcycle_verified.ids.labels` & `lucapcycle_verified.fasta`: checked predicted positives by LucaPCycle.
`lucapcycle_unverifiable.ids` & `lucapcycle_unverifiable.ids`: unverifiable predicted positives by LucaPCycle.
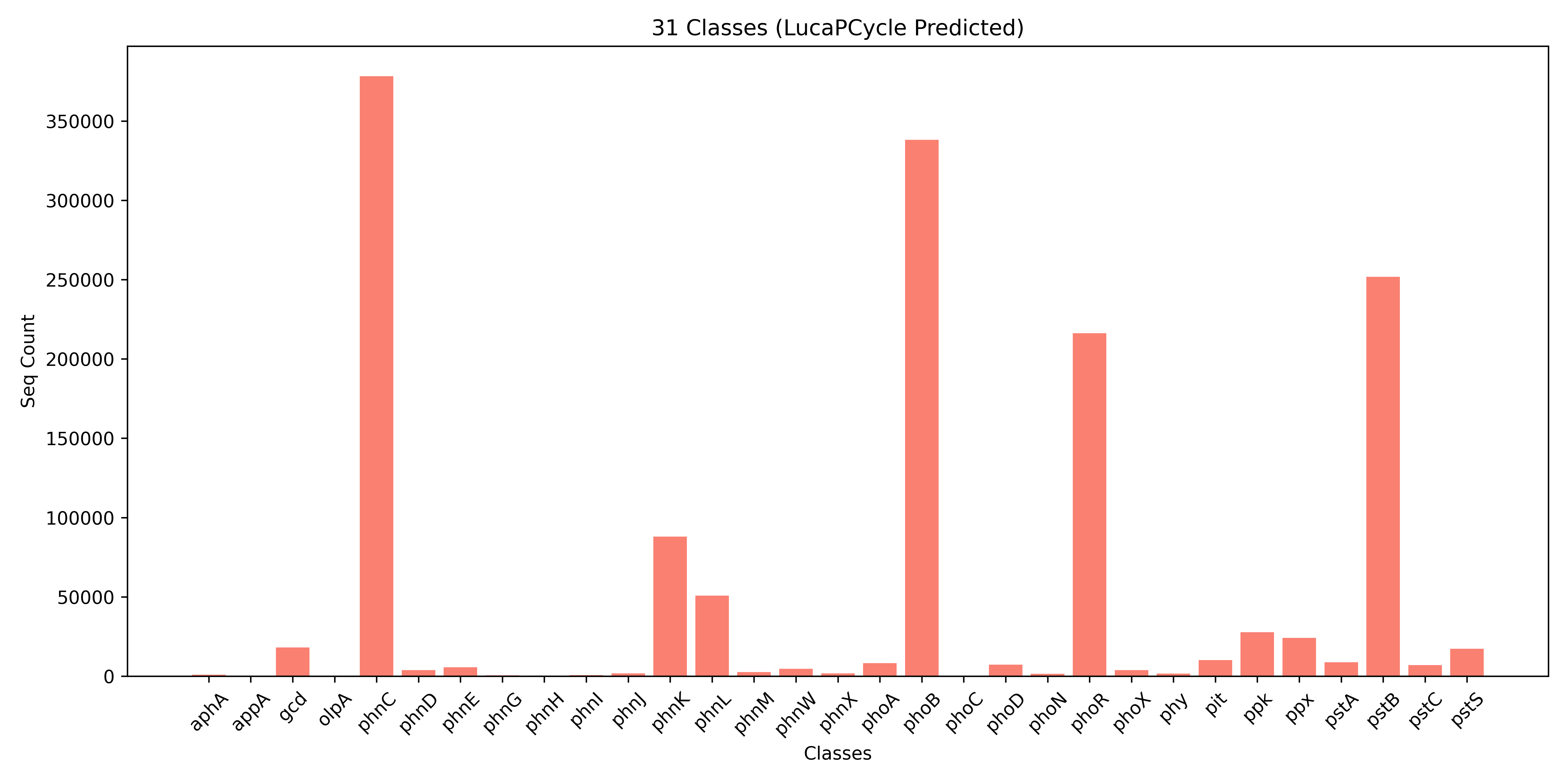 **Fig.2 The Predicted Details.**
* **Diamond Blastp**
Results in **`results/Blastp/`** or Blastp Results FTP
`blastp_init.ids.labels` & `blastp_init.fasta`: unchecked predicted positives by Blastp.
`blastp_verified.ids.labels` & `blastp_verified.fasta`: checked predicted positives by Blastp.
* **KofamScan**
Results in **`results/KofamScan/`** or KofamScan FTP
`kofamscan_init.ids.labels` & `kofamscan_init.fasta`: unchecked predicted positives by KofamScan.
`kofamscan_verified.ids.labels` & `kofamscan_verified.fasta`: checked predicted positives by KofamScan.
**Fig.2 The Predicted Details.**
* **Diamond Blastp**
Results in **`results/Blastp/`** or Blastp Results FTP
`blastp_init.ids.labels` & `blastp_init.fasta`: unchecked predicted positives by Blastp.
`blastp_verified.ids.labels` & `blastp_verified.fasta`: checked predicted positives by Blastp.
* **KofamScan**
Results in **`results/KofamScan/`** or KofamScan FTP
`kofamscan_init.ids.labels` & `kofamscan_init.fasta`: unchecked predicted positives by KofamScan.
`kofamscan_verified.ids.labels` & `kofamscan_verified.fasta`: checked predicted positives by KofamScan.
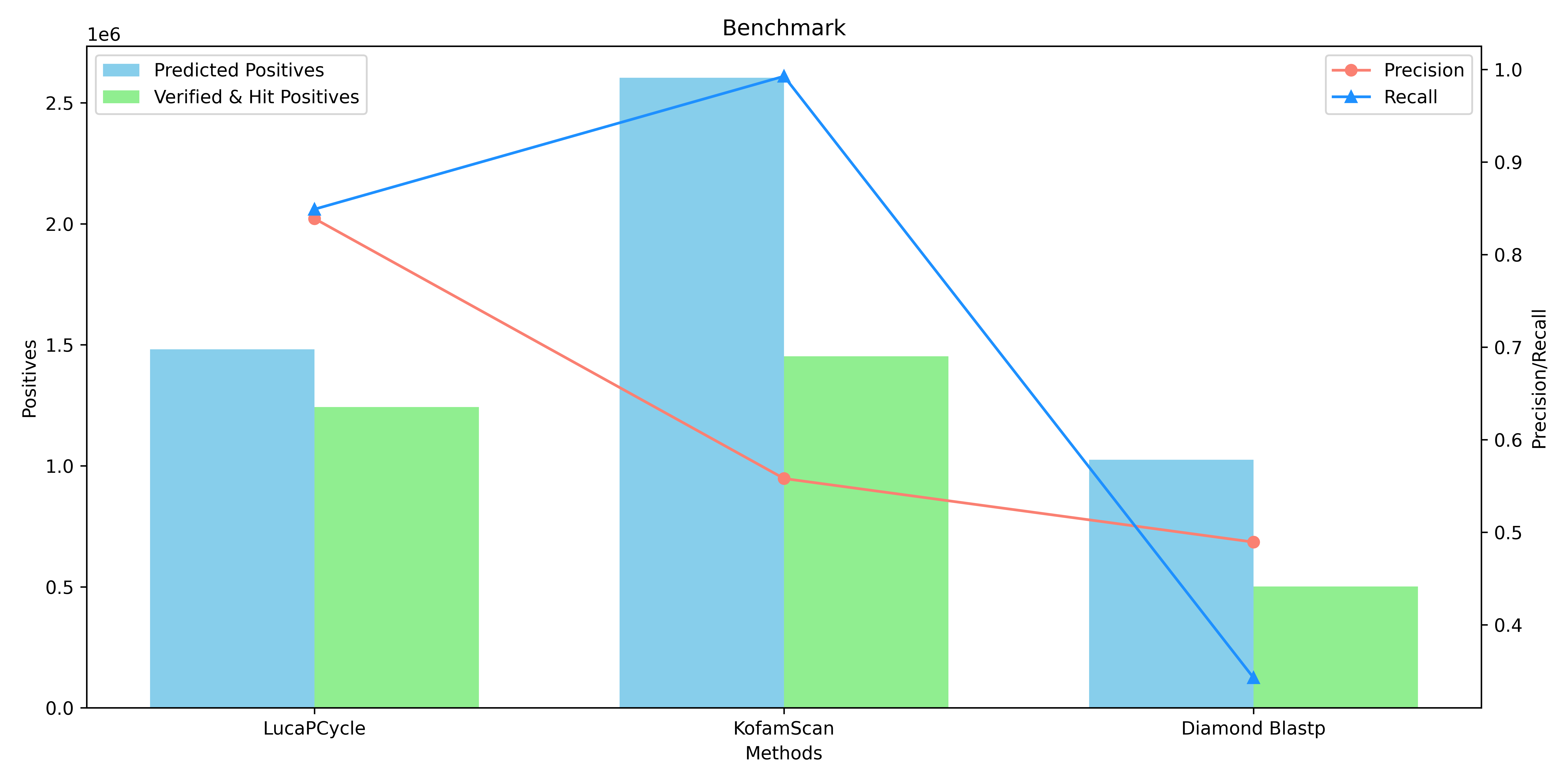 **Fig.3 Benchmark.**
### 3) Tree-Families
Sequence Tree
Phylogenetic tree of alkaline phosphatase with remote homology based on protein sequences.
Structural Tree
Structure-based phylogeny of alkaline phosphatase with remote homology and reference proteins.
Families
Representatives from non-singleton P-solubilizing protein families.
## 7. Contributor
Yong He,
Zhaorong Li,
Chuwen Zhang,
Xiyang Dong
## 8. Citation
@article {
Zhang2024.07.09.602434,
author = {Zhang, Chuwen and He, Yong and Wang, Jieni and Chen, Tengkai and Baltar, Federico and Hu, Minjie and Liao, Jing and Xiao, Xi and Li, Zhao-Rong and Dong, Xiyang},
title = {Illuminating microbial phosphorus cycling in deep-sea cold seep sediments using protein language models},
elocation-id = {2024.07.09.602434},
year = {2024},
doi = {10.1101/2024.07.09.602434},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/07/09/2024.07.09.602434},
eprint = {https://www.biorxiv.org/content/early/2024/07/09/2024.07.09.602434.full.pdf},
journal = {bioRxiv}
}
**Fig.3 Benchmark.**
### 3) Tree-Families
Sequence Tree
Phylogenetic tree of alkaline phosphatase with remote homology based on protein sequences.
Structural Tree
Structure-based phylogeny of alkaline phosphatase with remote homology and reference proteins.
Families
Representatives from non-singleton P-solubilizing protein families.
## 7. Contributor
Yong He,
Zhaorong Li,
Chuwen Zhang,
Xiyang Dong
## 8. Citation
@article {
Zhang2024.07.09.602434,
author = {Zhang, Chuwen and He, Yong and Wang, Jieni and Chen, Tengkai and Baltar, Federico and Hu, Minjie and Liao, Jing and Xiao, Xi and Li, Zhao-Rong and Dong, Xiyang},
title = {Illuminating microbial phosphorus cycling in deep-sea cold seep sediments using protein language models},
elocation-id = {2024.07.09.602434},
year = {2024},
doi = {10.1101/2024.07.09.602434},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/07/09/2024.07.09.602434},
eprint = {https://www.biorxiv.org/content/early/2024/07/09/2024.07.09.602434.full.pdf},
journal = {bioRxiv}
}
 **Fig.1 LucaPCycle.**
**Fig.1 LucaPCycle.**
 **Fig.1 LucaPCycle.**
**Fig.1 LucaPCycle.**
 **Fig.2 The Predicted Details.**
**Fig.2 The Predicted Details.**
 **Fig.3 Benchmark.**
**Fig.3 Benchmark.**